- 科研进展
-
刘全中副教授在基于深度学习的DNA甲基化位点预测方法取得新进展
作者: 来源: 发布日期:2020-11-25 浏览次数:论文题目:DeepTorrent: a deep learning-based approach for predicting DNA N4-methylcytosine sites
作 者:Quanzhong Liu, Jinxiang Chen, Yanze Wang, Shuqin Li, Cangzhi Jia, Jiangning Song and Fuyi Li
期刊名称:Briefings in Bioinformatics(CCF B类)
发表时间:2020年7月
论文摘要:
DNA N4-methylcytosine (4mC) is an important epigenetic modification that plays a vital role in regulating DNA replication and expression. However, it is challenging to detect 4mC sites through experimental methods, which are time-consuming and costly. Thus, computational tools that can identify 4mC sites would be very useful for understanding the mechanism of this important type of DNA modification. Several machine learning-based 4mC predictors have been proposed in the past 3 years, although their performance is unsatisfactory. Deep learning is a promising technique for the development of more accurate 4mC site predictions. In this work, we propose a deep learning-based approach, called DeepTorrent, for improved prediction of 4mC sites from DNA sequences. It combines four different feature encoding schemes to encode raw DNA sequences and employs multi-layer convolutional neural networks with an inception module integrated with bidirectional long short-term memory to effectively learn the higher-order feature representations. Dimension reduction and concatenated feature maps from the filters of different sizes are then applied to the inception module. In addition, an attention mechanism and transfer learning techniques are also employed to train the robust predictor. Extensive benchmarking experiments demonstrate that DeepTorrent significantly improves the performance of 4mC site prediction compared with several state-of-the-art methods.
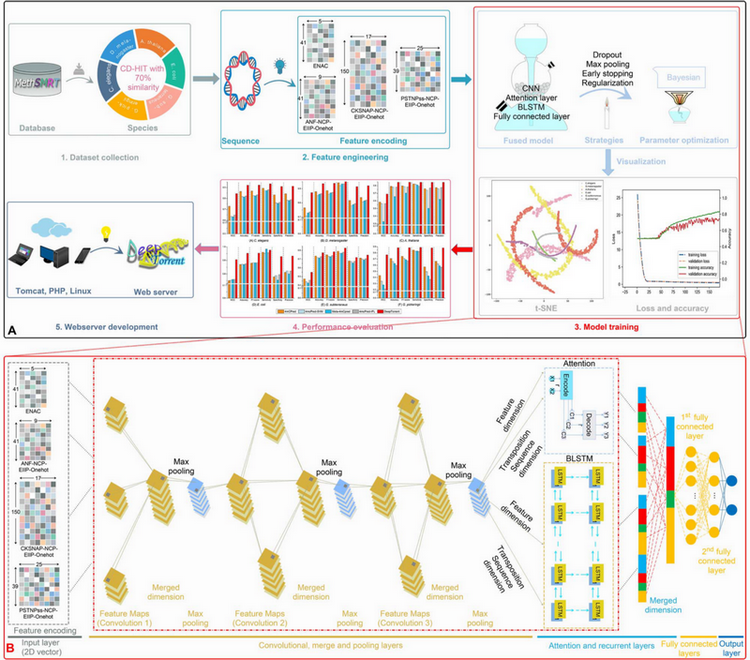
Webserver and source code: http://DeepTorrent.erc.monash.edu/